Genetics of Autism Spectrum Disorders, Dr. Abha Gupta – Video
Genetics of Autism Spectrum Disorders, Dr. Abha Gupta
Dr. Gupta discusses the evidence for a genetic etiology in autism and the results of various genetics studies of autism. She also reviews recommendations for clinical genetics testing and future...
By: YaleUniversity
Read this article:
Genetics of Autism Spectrum Disorders, Dr. Abha Gupta - Video
Recommendation and review posted by Bethany Smith
The Sims 3 | Perfect Genetics Challenge Part 16: Cotton Candy #3? – Video
The Sims 3 | Perfect Genetics Challenge Part 16: Cotton Candy #3?
In this part, i introduce you to our new baby to the family! Only some people will understand the title 😉 Backstory: "Once upon a time, the Mighty Player sent a Sim to live in the world...
By: simplyapril
Visit link:
The Sims 3 | Perfect Genetics Challenge Part 16: Cotton Candy #3? - Video
Recommendation and review posted by Bethany Smith
Genetics – The Cult (prod by. Besmorpheous) – Video
Genetics - The Cult (prod by. Besmorpheous)
Download our mixtape in the link below http://genetics4.bandcamp.com/album/aggressive-expansion play in 1080p for best quality.
By: Genetics
Continued here:
Genetics - The Cult (prod by. Besmorpheous) - Video
Recommendation and review posted by Bethany Smith
Minecraft Mod Sauce Ep. 6 – Flight & Advanced Genetics ( HermitCraft Modded Minecraft ) – Video
Minecraft Mod Sauce Ep. 6 - Flight Advanced Genetics ( HermitCraft Modded Minecraft )
Minecraft Modded Minecraft Mod Sauce Hermitcraft More ModSauce: https://www.youtube.com/playlist?list=PL7vFECXWtNMEOEGuR8jCkB93DWdxejW6Y Please "Like" this video if you want more !!!...
By: KingDaddyDMAC
Read the original here:
Minecraft Mod Sauce Ep. 6 - Flight & Advanced Genetics ( HermitCraft Modded Minecraft ) - Video
Recommendation and review posted by Bethany Smith
Race and Genetics – Video
Race and Genetics
Biotechnology is racing forward, perpetuating the idea that human variation can be neatly packaged in old stereotypes and racial categorizations.
By: TheCRGChannel2
Read the original:
Race and Genetics - Video
Recommendation and review posted by Bethany Smith
'Green card' for beef genetics
THE export of Australian cattle genetics has the potential to earn $3.5 million a year - and it just moved one step closer.
Under a new agreement between Australia and the US, seedstock producers will no longer have to carry out expensive tuberculosis and bluetongue testing, a move described by many as a trade barrier.
Minister of Agriculture Barnaby Joyce made the announcement earlier this week, and although the US was yet to recognise Australia's TB-free status, seedstock producers were hailing the development as a major breakthrough.
"There is the potential for northern Australia's cattle studs especially to increase their exports significantly because of these changes," Mr Joyce said.
Artificial breeding expert and veterinarian Ced Wise has welcomed the move and said although the US market was relatively low volume, it had a high value.
It currently accounts for more than half the value of all cattle semen and embryo exports from Australia.
"For those select producers who are producing genetics of the highest order, I think it will be an important market and a valuable addition," he said.
While lauding the move, Dr Wise said more work was needed. "We still have a few hurdles to jump, but they are at least 'jumpable' and they can be done for that market."
While the Department of Agriculture has given with one hand, it has taken with the other, as its fees for processing increased from $283 an hour to $954 an hour from July 1.
This has since decreased to roughly $400 an hour, Dr Wise said.
Read more from the original source:
'Green card' for beef genetics
Recommendation and review posted by Bethany Smith
Gene Therapy, Trials in Down Syndrome by Siham Al-Lawati – Video
Gene Therapy, Trials in Down Syndrome by Siham Al-Lawati
Gene Therapy, Trials in Down Syndrome.
By: National Genetic Centre /
Read more from the original source:
Gene Therapy, Trials in Down Syndrome by Siham Al-Lawati - Video
Recommendation and review posted by Bethany Smith
What does personalized medicine mean to you and your family? – Video
What does personalized medicine mean to you and your family?
In June 2014, our gift club members attended a special panel discussion featuring Drs. Patricia Dawson, Hank Kaplan, Tanya Wahl of Swedish Cancer Institute (...
By: SwedishFoundation
See the rest here:
What does personalized medicine mean to you and your family? - Video
Recommendation and review posted by sam
Team reveals molecular competition drives adult stem cells to specialize
1 hour ago A bam mutant fruit fly ovary, known as the germanium, contains only adult stem cell-like cells (red) and spherical spectrosome (green). The accumulation of only adult stem cell-like cells indicates a mutation in the master differentiation factor bam completely blocks germline stem cell lineage differentiation. Credit: Ting Xie, Ph.D., Stowers Institute for Medical Research
Adult organisms ranging from fruit flies to humans harbor adult stem cells, some of which renew themselves through cell division while others differentiate into the specialized cells needed to replace worn-out or damaged organs and tissues.
Understanding the molecular mechanisms that control the balance between self-renewal and differentiation in adult stem cells is an important foundation for developing therapies to regenerate diseased, injured or aged tissue.
In the current issue of the journal Nature, scientists at the Stowers Institute for Medical Research report that competition between two proteins, Bam and COP9, balances the self-renewal and differentiation functions of ovarian germline stem cells (GSCs) in fruit flies (Drosophila melanogaster).
"Bam is the master differentiation factor in the Drosophila female GSC system," says Stowers Investigator Ting Xie, Ph.D., and senior author of the Nature paper. "In order to carry out the switch from self-renewal to differentiation, Bam must inactivate the functions of self-renewing factors as well as activate the functions of differentiation factors."
Bam, which is encoded by the gene with the unusual name of bag-of-marbles, is expressed at high levels in differentiating cells and very low levels in GSCs of fruit flies.
Among the self-renewing factors targeted by Bam is the COP9 signalosome (CSN), an evolutionarily conserved, multi-functional complex that contains eight protein sub-units (CSN1 to CSN8). Xie and his collaborators discovered that Bam and the COP9 sub-unit known as CSN4 have opposite functions in regulating the fate of GSCs in female fruit flies.
Bam can switch COP9 function from self-renewal to differentiation by sequestering and antagonizing CSN4, Xie says. "Bam directly binds to CSN4, preventing its association with the seven other COP9 components via protein competition," he adds. CSN4 is the only COP9 sub-unit that can interact with Bam.
"This study has offered a novel way for Bam to carry out the switch from self-renewal to differentiation," says Xie, whose lab uses a combination of genetic, molecular, genomic and cell biological approaches to investigate GSCs as well as somatic stem cells of fruit flies.
In the Nature paper, Xie's lab also reports that CSN4 is the only one of the eight sub-units that is not involved in the regulation of GSC differentiation of female fruit flies. "One possible explanation for the opposite effects of CSN4 and the other CSN proteins is that the sequestration of CSN4 by Bam allows the other CSN proteins to have differentiation-promoting functions," he says.
Continue reading here:
Team reveals molecular competition drives adult stem cells to specialize
Recommendation and review posted by Bethany Smith
Researchers seek 'safety lock' against tumor growth after stem cell transplantation
PUBLIC RELEASE DATE:
6-Aug-2014
Contact: Robert Miranda cogcomm@aol.com Cell Transplantation Center of Excellence for Aging and Brain Repair
Putnam Valley, NY. (Aug. 6, 2014) Recent studies have shown that transplanting induced pluripotent stem cell-derived neural stem cells (iPS-NSCs) can promote functional recovery after spinal cord injury in rodents and non-human primates. However, a serious drawback to the transplantation of iPS-NSCs is the potential for tumor growth, or tumorogenesis, post-transplantation.
In an effort to better understand this risk and find ways to prevent it, a team of Japanese researchers has completed a study in which they transplanted a human glioblastoma cell line into the intact spinal columns of laboratory mice that were either immunodeficient or immunocompetent and treated with or without immunosuppresant drugs. Bioluminescent imaging was used to track the transplanted cells as they were manipulated by immunorejection.
The researchers found that the withdrawal of immunosuppressant drugs eliminated tumor growth and, in effect, created a 'safety lock' against tumor formation as an adverse outcome of cell transplantation. They also confirmed that withdrawal of immunosuppression led to rejection of tumors formed by transplantation of induced pluripotent stem cell derived neural stem/progenitor cells (iPS-NP/SCs).
Although the central nervous system has shown difficulty in regenerating after damage, transplanting neural stem/progenitor cells (NS/PCs) has shown promise. Yet the problem of tumorogenesis, and increases in teratomas and gliomas after transplantation has been a serious problem. However, this study provides a provisional link to immune therapy that accompanies cell transplantation and the possibility that inducing immunorejection may work to reduce the likelihood of tumorogenesis occurring.
"Our findings suggest that it is possible to induce immunorejection of any type of foreign-grafted tumor cells by immunomodulation," said study co-author Dr. Masaya Nakamura of the Keio University School of Medicine. "However, the tumorogenic mechanisms of induced pluripotent neural stem/progenitor cells (iPS-NS/PCs) are still to be elucidated, and there may be differences between iPS-NS/PCs derived tumors and glioblastoma arising from genetic mutations, abnormal epigenetic modifications and altered cell metabolisms."
The researchers concluded that their model might be a reliable tool to target human spinal cord tumors in preclinical studies and also useful for studying the therapeutic effect of anticancer drugs against malignant tumors.
"This study provides evidence that the use of, and subsequent removal of, immunosuppression can be used to modulate cell survival and potentially remove tumor formation by transplanted glioma cells and provides preliminary data that the same is true for iPS-NS/PCs." said Dr. Paul Sanberg, distinguished professor at the Center of Excellence for Aging and Brain Repair, University of South Florida. "Further study is required to determine if this technique could be used under all circumstances where transplantation of cells can result in tumor formation and its reliability in other organisms and paradigms."
Visit link:
Researchers seek 'safety lock' against tumor growth after stem cell transplantation
Recommendation and review posted by Bethany Smith
Sound Advice Concerning How To Stay Young
morrisrodney594 posted a photo:
Getting Older can not be avoided by anyone. While many people carry it in stride and weather growing older well, others have a problem with it. Using easy ideas, it is possible to slow growing older, and feel younger well to your later years.
Encourage the mind to be active through learning. Aging is normally intermingled with intelligence you ought to increase your intelligence up to you can actually. Everything that energizes the brain, say for example a computer course or maybe a crossword puzzle, helps keep the mind active, so you feeling in addition to your game.
Aging can be difficult. There comes a time in many peoples lives when they must depend on others to take care of them because they can no longer do it themselves. This would be the time that you need to consider moving into a nursing home. Although many people are resistant to living in a nursing home, you can find situations where this is the best choice for everyone involved. People that work in these places will be able to give you the health care you are unable to allow yourself.
Having good friends is a great way to make yourself feel great. You happen to be never too old to create new friendships. Meet new people by strike up conversations with strangers in the grocery line, on the bus or in an event. It will improve your life.
Personalize your home to make it your own. You need to adjust your home to your preferences when you age. When moving to a new home, make sure you decorate your home with items that give you comfort and make you feel welcome.
Proper rest is essential to good health. Sleeping seven or nine hours can both relax you and also allow you to retain a proper hormonal balance. Running on inadequate sleep also causes you to grouchy and annoying to become around.
If you are getting older, hormone balance will be an essential alternation in you life. Whenever your hormones are unbalanced, you may are afflicted by insomnia as well as putting on weight. As you age, these can result in further problems. Talk to your physician to find the best advice regarding how to control hormone imbalances. This could benefit your later years in tremendous ways.
Reading the recommendation we now have offered has armed you together with the appropriate knowledge to stall the entire process of aging. Better of luck in staying youthful! personal-longevity.com/2014/07/27/gene-that-plays-a-surpr...

See more here:
Sound Advice Concerning How To Stay Young
Recommendation and review posted by Bethany Smith
Personalized Medicine and Companion Diagnostics Go Hand-in-Hand
The U.S. Food and Drug Administration posted a photo:
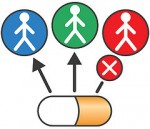
Companion diagnostic tests show which patients could be helped by a drug and which patients would not benefit, and could even be harmed.
The recent approval of a genetic test to help doctors prescribe a drug that treats colorectal cancer is just one example of the increasing importance of companion diagnostic tests in personalized medicine to ensure the safety and effectiveness of targeted therapies. Read this FDA Consumer Update to learn more.
This graphic is free of all copyright restrictions and available for use and redistribution without permission. Credit to the U.S. Food and Drug Administration is appreciated but not required. For more privacy and use information visit: http://www.flickr.com/people/fdaphotos/
FDA graphic by Michael J. Ermarth
Read the rest here:
Personalized Medicine and Companion Diagnostics Go Hand-in-Hand
Recommendation and review posted by Bethany Smith
Amber Wave of Grain
Todd Klassy posted a photo:
The sun shines down on a field of wheat ready for harvest near Shonkin, Montana.
? my blog
? my facebook
? my twitter
? my website
? my youtube
? my e-mail
© 2010 Todd Klassy. All Rights Reserved.

See the original post here:
Amber Wave of Grain
Recommendation and review posted by Bethany Smith
New standards proposed for reporting spinal cord injury experiments
PUBLIC RELEASE DATE:
6-Aug-2014
Contact: Kathryn Ryan kryan@liebertpub.com 914-740-2100 Mary Ann Liebert, Inc./Genetic Engineering News
New Rochelle, NY, August 6, 2014The difficulty in replicating and directly comparing and confirming the scientific results reported by researchers worldwide who are studying new approaches to treating spinal cord injuries is slowing the translation of important new findings to patient care. A newly proposed reporting standard for spinal cord injury (SCI) experimentation defines the minimum information that is appropriate for modeling an SCI in the research setting, as presented in an article in Journal of Neurotrauma, a peer-reviewed publication from Mary Ann Liebert, Inc., publishers. The article is available Open Access on the Journal of Neurotrauma website.
In the article, "Minimum Information about a Spinal Cord Injury Experiment: A Proposed Reporting Standard for Spinal Cord Injury Experiments" Vance P. Lemmon and a team of coauthors from University of Miami School of Medicine (Florida), University of California San Francisco, The Ohio State University (Columbus), Indiana University (Indianapolis), University of Kentucky (Lexington), and Niigata University (Japan), representing the MIASCI Consortium, describe how the adoption of uniform reporting standards and the use of common data elements can improve transparency in scientific reporting and facilitate the development of databases of experimental information"computer-readable knowledge repositories."
"This manuscript from many of the leading researchers in the field of spinal cord research should provide uniform databases for researchers to review new findings in this rapidly growing field and promote the successful translation of treatments to the clinic," says W. Dalton Dietrich, PhD, Deputy Editor of Journal of Neurotrauma and Kinetic Concepts Distinguished Chair in Neurosurgery, Professor of Neurological Surgery, Neurology and Cell Biology, University of Miami Leonard M. Miller School of Medicine.
###
About the Journal
Journal of Neurotrauma is an authoritative peer-reviewed journal published 24 times per year in print and online that focuses on the latest advances in the clinical and laboratory investigation of traumatic brain and spinal cord injury. Emphasis is on the basic pathobiology of injury to the nervous system, and the papers and reviews evaluate preclinical and clinical trials targeted at improving the early management and long-term care and recovery of patients with traumatic brain injury. Journal of Neurotrauma is the official journal of the National Neurotrauma Society and the International Neurotrauma Society. Complete tables of content and a sample issue may be viewed on the Journal of Neurotrauma website.
About the Publisher
Excerpt from:
New standards proposed for reporting spinal cord injury experiments
Recommendation and review posted by Bethany Smith
Zoe 6 months after she was a quadriplegic from spinal cord injury – Video
Zoe 6 months after she was a quadriplegic from spinal cord injury
By: Paint Me Riding
See original here:
Zoe 6 months after she was a quadriplegic from spinal cord injury - Video
Recommendation and review posted by sam
Blood Pressure Problems After a Spinal cord injury – Video
Blood Pressure Problems After a Spinal cord injury
Autonimic dysreflexia occurs after a spinal cord injury. It can cause a heart attack or a stroke if blood pressure is not properly managed. If you wish to kn...
By: CloStar Rose
Excerpt from:
Blood Pressure Problems After a Spinal cord injury - Video
Recommendation and review posted by sam
Scar Removal Orlando | Cicatrices | Eterna MD – Video
Scar Removal Orlando | Cicatrices | Eterna MD
Watch the scar removal procedure performed by Dr. Carlos Mercado of Eterna MD. Learn more by visiting our website at http://eternamd.com.
By: Eterna MD Regenerative Medicine
More here:
Scar Removal Orlando | Cicatrices | Eterna MD - Video
Recommendation and review posted by sam
Yoshiki Sasai Suicide: Japanese Stem Cell Scientist Found Dead In Kobe Facility
A Japanese scientist who was among a team of researchers accused of falsifying the results of two stem cell studies committed suicide Tuesday at a government science institute in western Japan. Yoshiki Sasai, deputy director of the Riken Center for Developmental Biology, was found by a security guard at the Kobe facility with a rope around his neck, the Associated Press reports. Authorities said he had suffered from cardiac arrest and was pronounced dead two hours later.
Sasai, 52, was considered an expert in embryonic stem cell research and co-authored two research papers published in January in the journal Nature that detailed a seemingly groundbreaking method of harvesting stem cells to grow new human tissue. Sasai and lead author Haruko Obokata reported having successfully altered ordinary mouse cells into versatile stem cells by immersing them in a mildly acidic solution. The resulting cells were named stimulus-triggered acquisition of pluripotency (STAP) cells.
The studies were initially praised as being on the cutting edge of stem cell treatment, but were quickly disputed when other scientists could not replicate the experimental procedure. The papers were retracted six months later after the journal found they contained erroneous data, among other flaws.
Scientists at RIKEN Center for Developmental Biology in Kobe are deeply concerned about the allegations regarding the recently reported STAP cells, the center said in a statement released in March. We wish to express our strong commitment to maintaining the highest level of scientific integrity to the public and the scientific community. We are fully aware that trust from the society is crucial for research activities carried out in RIKEN.
The scandal apparently affected Sasais health. Following the initial revelation that the research he was involved in may have been flubbed, he was hospitalized in March for stress, according to Riken spokesman Satoru Kagaya, who told reporters during a televised news conference on Tuesday that Sasai "seemed completely exhausted" when they talked over the phone in May.
Several suicide notes were found on Sasais secretarys desk, according to the Wall Street Journal. The content of the notes has not been made public, but officials said two of the notes were addressed to Riken officials, one of whom was Obokata.
Link:
Yoshiki Sasai Suicide: Japanese Stem Cell Scientist Found Dead In Kobe Facility
Recommendation and review posted by Bethany Smith
Japanese scientist stem-cell scientist Yoshiki Sasai commits suicide
Yoshiki Sasai, who was embroiled in a stem-cell scandal, committed suicide He was found with a rope around his neck at science institute Riken in Japan Mr Sasai, 52, was deputy chief of Riken's Center for Developmental Biology He co-authored stem-cell research papers with falsified contents
By Ted Thornhill
Published: 06:20 EST, 5 August 2014 | Updated: 13:25 EST, 5 August 2014
A senior Japanese scientist embroiled in a stem-cell research scandal died on Tuesday in an apparent suicide, police said.
Yoshiki Sasai, who supervised and co-authored stem-cell research papers that had to be retracted due to falsified contents, was found suffering from cardiac arrest at the government-affiliated science institute Riken in Kobe, in western Japan, according to Hyogo prefectural police.
Sasai, 52, was deputy chief of Riken's Center for Developmental Biology.
Scroll down for video
Tragic:Yoshiki Sasai, who was embroiled in a stem-cell scandal, committed suicide and was found with a rope around his neck at his place of work
A security guard found him with a rope around his neck, according to Riken. Sasai was rushed to a hospital, but was pronounced dead two hours later.
Police and Riken said Sasai left what appeared to be suicide notes, but refused to disclose their contents.
Read more:
Japanese scientist stem-cell scientist Yoshiki Sasai commits suicide
Recommendation and review posted by Bethany Smith
Implanted brain cells integrate fully with mouse brain tissue
Brain cells that were grafted into the brains of mice have become fully functionally integrated after six months. The successful neuron transplant could pave the way for therapies to treat neurodegenerative diseases such as Parkinson's.
A team of stem cell researchers at the Luxembourg Centre for Systems Biomedicine created the grafted neurons -- induced neuronal stem cells -- in a petri dish out of the host's reprogrammed skin cells. This technique dramatically improved the compatibility of the implanted cells.
Six months after the brain cells were implanted into the hippocampus and cortex regions of the brain, the neurons were fully integrated with the original brain cells via newly formed synapses (the contact points between neurons). The induced neuronal stem cells had changed into different types of brain cells -- neurons, astrocytes and oligodendrocytes -- over time within the host brain. Functional integration with the existing network of cells is absolutely critical for long-term survival of the new brain tissue. The new brain cells exhibited normal activity in tests and the mice showed no adverse side effects.
The plan for researchers is now to explore replacing the type of neurons that tend to die off in the brain of Parkinson's patients -- those neurons found in the substantia nigra that produce dopamine. It may, in the future, be possible to implant neurons to produce the diminished dopamine, which could prove to be an effective treatment for the disease.
Of course, it's a bit leap from the current research to human trials. "Successes in human therapy are still a long way off, but I am sure successful cell replacement therapies will exist in future," says team leader and stem cell researcher Jens Schwamborn. "Our research results have taken us a step further in this direction."
The study has been published in Stem Cell Reports and is available to read for free.
The rest is here:
Implanted brain cells integrate fully with mouse brain tissue
Recommendation and review posted by Bethany Smith
Implanted neurons become part of the brain, mouse study shows
Scientists at the Luxembourg Centre for Systems Biomedicine (LCSB) of the University of Luxembourg have grafted neurons reprogrammed from skin cells into the brains of mice for the first time with long-term stability. Six months after implantation, the neurons had become fully functionally integrated into the brain. This successful, lastingly stable, implantation of neurons raises hope for future therapies that will replace sick neurons with healthy ones in the brains of Parkinson's disease patients, for example.
The Luxembourg researchers published their results in the current issue of Stem Cell Reports.
The LCSB research group around Prof. Dr. Jens Schwamborn and Kathrin Hemmer is working continuously to bring cell replacement therapy to maturity as a treatment for neurodegenerative diseases. Sick and dead neurons in the brain can be replaced with new cells. This could one day cure disorders such as Parkinson's disease. The path towards successful therapy in humans, however, is long. "Successes in human therapy are still a long way off, but I am sure successful cell replacement therapies will exist in future. Our research results have taken us a step further in this direction," declares stem cell researcher Prof. Schwamborn, who heads a group of 15 scientists at LCSB.
In their latest tests, the research group and colleagues from the Max Planck Institute and the University Hospital Mnster and the University of Bielefeld succeeded in creating stable nerve tissue in the brain from neurons that had been reprogrammed from skin cells. The stem cell researchers' technique of producing neurons, or more specifically induced neuronal stem cells (iNSC), in a petri dish from the host's own skin cells considerably improves the compatibility of the implanted cells. The treated mice showed no adverse side effects even six months after implantation into the hippocampus and cortex regions of the brain. In fact it was quite the opposite -- the implanted neurons were fully integrated into the complex network of the brain. The neurons exhibited normal activity and were connected to the original brain cells via newly formed synapses, the contact points between nerve cells.
The tests demonstrate that the scientists are continually gaining a better understanding of how to treat such cells in order to successfully replace damaged or dead tissue. "Building upon the current insights, we will now be looking specifically at the type of neurons that die off in the brain of Parkinson's patients -- namely the dopamine-producing neurons," Schwamborn reports. In future, implanted neurons could produce the lacking dopamine directly in the patient's brain and transport it to the appropriate sites. This could result in an actual cure, as has so far been impossible. The first trials in mice are in progress at the LCSB laboratories on the university campus Belval.
Story Source:
The above story is based on materials provided by Universit du Luxembourg. Note: Materials may be edited for content and length.
Visit link:
Implanted neurons become part of the brain, mouse study shows
Recommendation and review posted by Bethany Smith
Luxury Skin Care: SkinStore.com Adds Reformulated, Repackaged DermaQuest
Gold River, CA (PRWEB) August 05, 2014
SkinStore.com, the nations leading e-commerce specialty retailer providing scientifically sound solutions for healing and maintaining healthy skin, has reintroduced DermaQuest to its assortment of premium products.
As the leader in botanical stem cell technology since 1999, DermaQuest is at the edge of innovation in advanced skincare. The luxurious formulas are rich in vitamins, peptides, plant stem cells and essential ingredients that hydrate, protect and actually rejuvenate the skin. Through specialized collections for every skin concern, and specific layering sequences to ensure maximum effectiveness and product absorption, DermaQuests formulas are able to realize the desires of any skin type. Their strict quality control and use of only superior ingredients has become unparalleled, results-oriented skincare: If they arent doing it, it simply cant be done yet.
The notable DermaQuest Stem Cell 3D Complex is powered by advanced Biotech Marine and botanical stem cells, peptides and potent antioxidants. The rich, silky formula was formulated to be a wonder tonic, a cure-all for the myriad signs of aging, such as fine lines, wrinkles, skin texture and tone.
Christina Bertolino, Senior, Buying Manager at SkinStore.com, said, DermaQuest offers the best of both worlds: luxury and proven results. The science behind the line is unparalleled and the visible effects speak for themselves.
About SkinStore.com. Physician-founded in 1997, SkinStore carries over 300 premium brands of skin care, cosmetics, hair care, beauty tools and fragrances from around the world, including high quality products normally found in luxury spas, fine department stores and dermatologist offices. An esthetician-staffed call center is available Monday through Friday to answer customer questions and help shoppers choose products best-suited for their skin type. The company is headquartered in Gold River (Sacramento), California. For more information visit SkinStore.com, SkincareStore.com.au or SkinStoreChina.com.
Contact Information Denise McDonald, Content & Production Manager SkinStore http://www.skinstore.com 916-475-1427
###
See more here:
Luxury Skin Care: SkinStore.com Adds Reformulated, Repackaged DermaQuest
Recommendation and review posted by Bethany Smith
Gene mutation may explain why some people need less sleep
Researchers have identified a gene mutation that may allow people who have it to function normally on less than six hours of sleep per night. People who have the gene also seem to be more resistant to the effects of sleep deprivation.
In a study on 100 pairs of twins, researchers found that a twin who had a variant of the BHLHE41 gene called p.Tyr362His slept for only about five hours per night, more than an hour shorter than his twin brother who did not carry the gene.
Moreover, the twin who carried the gene mutation had 40 percent fewer average lapses of performance during 38 hours without sleep, and required less recovery sleep after the period of sleep deprivation. The twin with the gene mutation only slept eight hours after being sleep-deprived for an extended period of time, compared with his brother, who slept for 9.5 hours.
7 Photos
These simple strategies from top sleep experts can help you fall asleep faster and sleep better
"This work provides an important second gene variant associated with sleep deprivation and for the first time shows the role of BHLHE41 in resistance to sleep deprivation in humans," study author Renata Pellegrino, a senior research associate in the Center for Applied Genomics at The Children's Hospital of Philadelphia, said in a statement. "The mutation was associated with resistance to the neurobehavioral effects of sleep deprivation."
All of the twins in the study were the same sex and were healthy. To conduct the study, the researchers measured the twins' sleep duration for seven to eight nights. The investigators also analyzed their response to 38 hours of sleep deprivation and length of recovery sleep. The twins' cognitive performance was examined every two hours during sleep deprivation.
The American Academy of Sleep Medicine recommends that adults get about seven to nine hours of sleep per night, even though individual sleep needs may vary. Very few people in the population are so-called natural short-sleepers who can get by on less than six hours of sleep without feeling tired and being less alert during the day.
But, according to the Centers for Disease Control and Prevention, 28 percent of U.S. adults report sleeping six hours or less in a 24-hour period. Many will suffer from daytime sleepiness and concentration problems as a result.
"This study emphasizes that our need for sleep is a biological requirement, not a personal preference," American Academy of Sleep Medicine President Dr. Timothy Morgenthaler, said in a statement. "Most adults appear to need at least seven hours of quality sleep each night for optimal health, productivity and daytime alertness."
Original post:
Gene mutation may explain why some people need less sleep
Recommendation and review posted by Bethany Smith
Gene editing world leaders boost Horizon SAB
Three of the worlds foremost experts in gene editing have joined the scientific advisory board of Cambridge UK medical technology innovator, Horizon Discovery.
CEO Dr Darrin Disley is hailing the appointments from Europe and Boston as a global triumph as the business supplies more and more research tools to accelerate progress towards personalised medicines.
The world renowned scientists who have joined Horizons SAB are Dr Emmanuelle Charpentier currently working in Germany and Drs Keith Joung and Feng Zhang from Massachusetts.
Dr Charpentier is head of the Department of Regulation in Infection Biology at the Helmholtz Centre for Infection Research in Braunschweig and Hannover Medical School. She is known worldwide for her work on bacterial immunity and uncovering key functional components of the bacterial CRISPR-Cas9 system.
Dr Charpentier was principal co-author on a seminal publication suggesting how the CRISPR-Cas9 system could be adapted from use in bacteria for broad application in mammalian genome editing. She is also a co-founder of ERS Genomics and CRISPR Therapeutics and was recently awarded the Alexander von Humboldt professorship and Dr Paul Janssen Award.
Dr Charpentier said: Horizon is fast becoming recognised as a leader in the field of gene editing, applying CRISPR and other technologies to develop innovative research for understanding the genetic basis of disease and the development of personalised medicines.
I am excited to help guide the next stage in the companys development and the application of CRISPR-Cas9 technology to their products and services.
Dr Joung has a long history in the development and application of engineered DNA-binding domains and systems for creating customised nucleases for genome editing. Over the past 10 years, his lab has pioneered methods for targeted genome editing using engineered zinc finger nucleases, transcription activator-like effector nucleases, as well as CRISPR-Cas9.
In a recent publication, Dr Joung and his co-workers described a novel system that combines CRISPR-Cas9 with a nuclease domain to generate dimeric RNA-guided FokI nucleases with greater specificity than first-generation CRISPR reagents.
Dr Joung is the recipient of a NIH Directors Pioneer Award and also a scientific co-founder of Editas Medicine. He currently serves as Associate Chief of Pathology for Research at Massachusetts General Hospital and is an Associate Professor of Pathology at Harvard Medical School.
See more here:
Gene editing world leaders boost Horizon SAB
Recommendation and review posted by Bethany Smith
Gene mutation makes people need less sleep
(CBS) - Researchers have identified a gene mutation that may allow people who have it to function normally on less than six hours of sleep per night. People who have the gene also seem to be more resistant to the effects of sleep deprivation.
In a study on 100 pairs of twins, researchers found that a twin who had a variant of the BHLHE41 gene called p.Tyr362His slept for only about five hours per night, more than an hour shorter than his twin brother who did not carry the gene.
Moreover, the twin who carried the gene mutation had 40 percent fewer average lapses of performance during 38 hours without sleep, and required less recovery sleep after the period of sleep deprivation. The twin with the gene mutation only slept eight hours after being sleep-deprived for an extended period of time, compared with his brother, who slept for 9.5 hours.
"This work provides an important second gene variant associated with sleep deprivation and for the first time shows the role of BHLHE41 in resistance to sleep deprivation in humans," study author Renata Pellegrino, a senior research associate in the Center for Applied Genomics at The Children's Hospital of Philadelphia, said in a statement. "The mutation was associated with resistance to the neurobehavioral effects of sleep deprivation."
All of the twins in the study were the same sex and were healthy. To conduct the study, the researchers measured the twins' sleep duration for seven to eight nights. The investigators also analyzed their response to 38 hours of sleep deprivation and length of recovery sleep. The twins' cognitive performance was examined every two hours during sleep deprivation.
The American Academy of Sleep Medicine recommends that adults get about seven to nine hours of sleep per night, even though individual sleep needs may vary. Very few people in the population are so-called natural short-sleepers who can get by on less than six hours of sleep without feeling tired and being less alert during the day.
But, according to the Centers for Disease Control and Prevention, 28 percent of U.S. adults report sleeping six hours or less in a 24-hour period. Many will suffer from daytime sleepiness and concentration problems as a result.
"This study emphasizes that our need for sleep is a biological requirement, not a personal preference," American Academy of Sleep Medicine President Dr. Timothy Morgenthaler, said in a statement. "Most adults appear to need at least seven hours of quality sleep each night for optimal health, productivity and daytime alertness."
The new study was published Monday in the journal Sleep.
See the rest here:
Gene mutation makes people need less sleep
Recommendation and review posted by Bethany Smith


